 Epitope Mapping at the Amino Acid Level
Epitope Mapping at the Amino Acid Level
Label-free screening of mAb libraries against a 384 peptide array.
The LSA can be used for antibody epitope discovery at the amino acid level by immobilizing a panel of up to 384 biotinylated peptides with overlapping sequences onto the SPR chip. A library of mAbs is then injected across the surface. The results can be processed with the Epitope Data Analysis Software to generate heatmap, dendrogram, and stacked plot data views to aid in selection.
Alternatively, enzymatic digestion can be used to generate different sequence contexts for epitope discovery, which can be immobilized on the array via flow printing and probed with antibody libraries or patient sera.
Epitope Mapping Heat Map
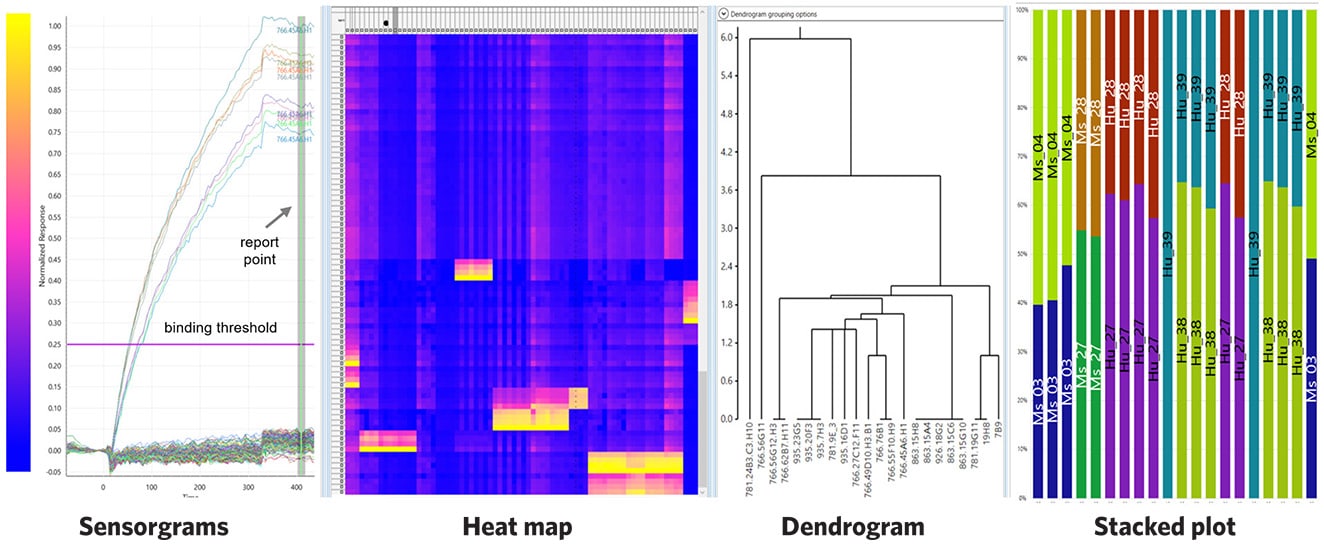
Eptitope Mapping on a 384-Peptide Array
Snapshot of the results of epitope mapping on a 384-peptide array showing sensorgrams, heat map, dendrogram, and stacked plot analyses.
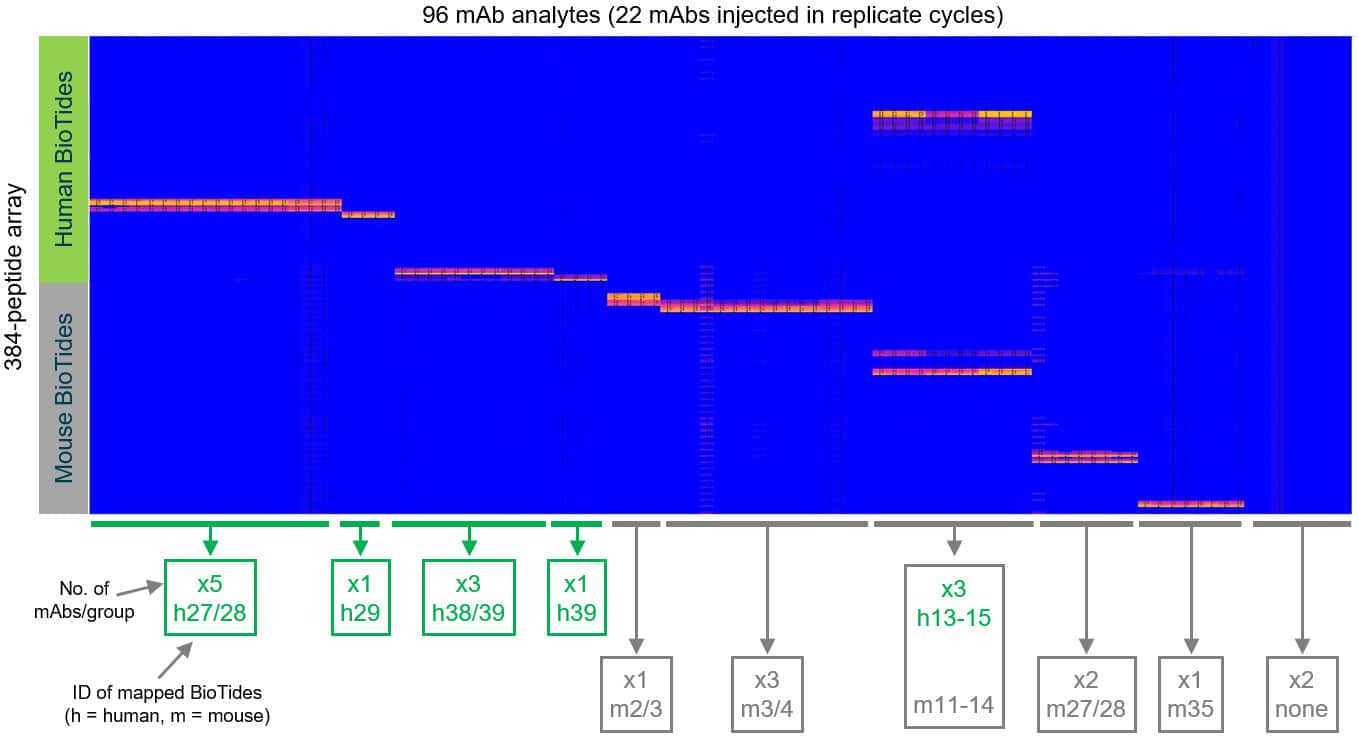
Sorted heat map showing how the mAbs cluster into different epitope groups based on their peptide specificities.
