Proximity-Based Degraders DNA Encoded Library (DEL) Compounds Kinase Inhibitors
Proximity-Based Degraders
Reimagine characterization of TPDs, PROTAC®s, LYTACs, AUTACs, RIBOTACs, and molecular glues using HT-SPR to gain truly meaningful throughput and multiplexing capabilities. One-on-many fluidics reduce sample quantities, consumable costs, and hands on time. Characterize binary and ternary kinetics for hundreds of interactions in parallel. Develop broad and nuanced views of cooperativity in the context of selectivity for up to hundreds of molecules in parallel.
- Throughput: Reduce assay run time and preserve activity of fragile targets/ligases
- Cooperativity: Be confident in the cooperativity values by inclusion of multiple replicates and controls
- Selectivity: Assess binding across up to hundreds of targets and off-targets simultaneously
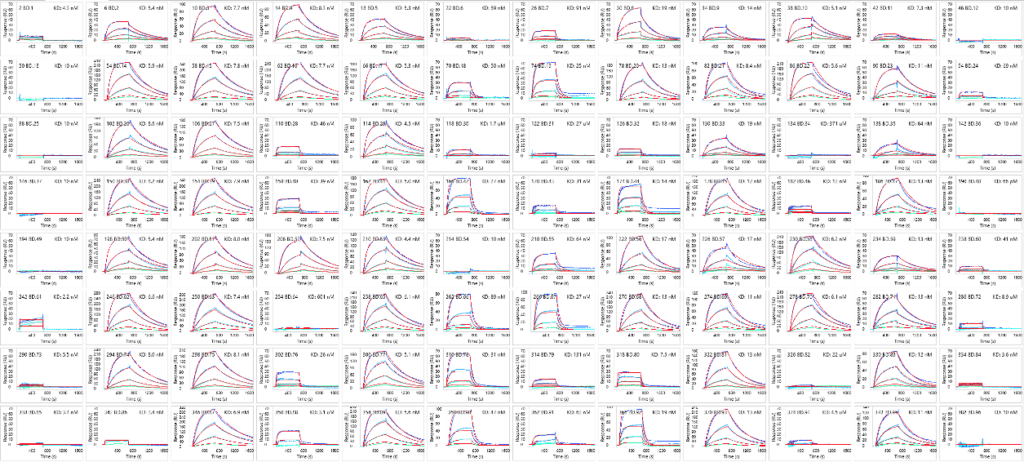
Ternary kinetic profiles of MZ1/VHL mixture binding to an array of 96 bromodomain proteins. 1:1 Langmuir fit lines in red.
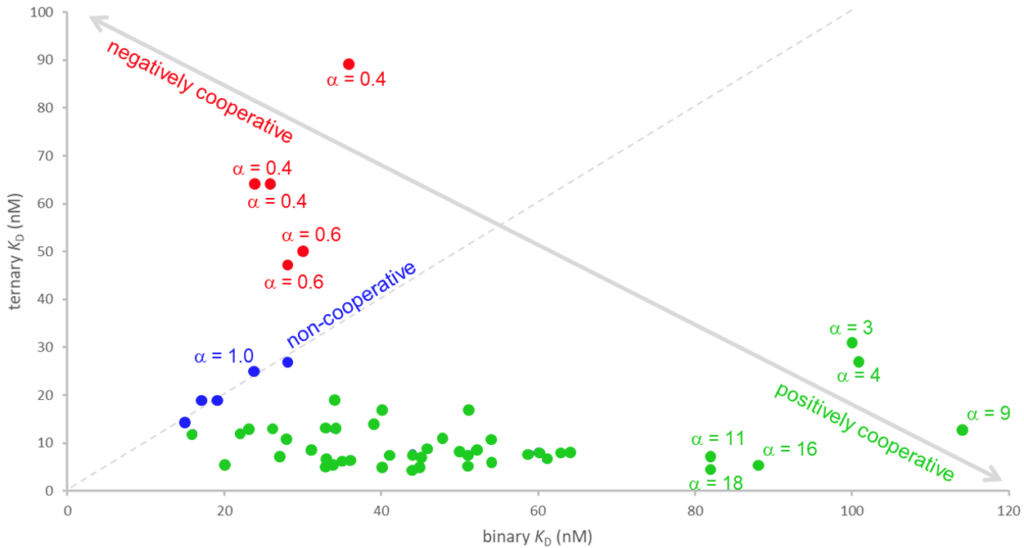
Cooperativity plot derived from binary and ternary affinities of bromodomain proteins in conjunction with MZ1 and VHL.
DNA Encoded Library (DEL) Compounds
Confidently determine detailed kinetic profiles (ka & kd) and affinity (KD) for DNA-tagged fragments, compounds, and targeted protein degraders (TPDs). Gold standard SPR technology affords real-time, label-free binding in exquisite detail. By leveraging the one-on-many power of the LSA®, arrays of 384 DELs can be screened in parallel with up to 1,152 in a single run.
- Data Quality: Highly resolved and accurate kinetics
- Speed: Characterize ≥ 5,700 compounds per week
- Simplicity: No need to resynthesize off-DNA and no challenges with compound solubility
- Flexibility: Potential to further increase throughput via combinatorial approaches
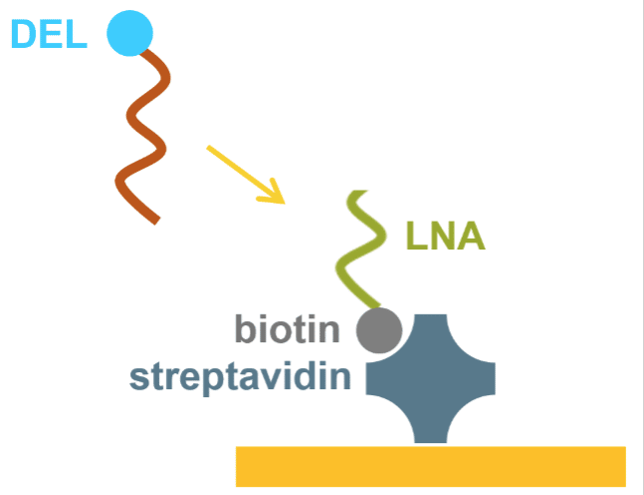
DNA-tagged fragments, compounds, or TPDs are arrayed as individual locations on a 384-spot array by oligonucleotide hybridization. Once binding kinetics against target and/or off-target are complete, the DNA-tagged species are easily denatured and removed from the surface allowing for another 384 molecules to be attached and characterized.

Detailed kinetic profiles showing well behaved binders (white), complex binders (purple), and non-binders (gray). Model fit lines are shown in red.
Kinase Inhibitors
Expand selectivity profiling to include up to hundreds of targets and off-targets in replicate in a single experiment. Enable detailed kinetic analysis across weak and high affinity binders in parallel. Distinguish subtle differences in selectivity with a high degree of accuracy. Throughput avoids the need to keep fragile kinases active for days of screening.
- Selectivity: Comprehensively screen candidates against hundreds of targets and off-targets
- Resolution: Quantitatively determine subtle differences in association and dissociation kinetics
- Broad dynamic range: Accurately determine kinetics for both weak and high affinity interactions in the same experiment
Selectivity of sunitinib against panel of kinases in duplicate.
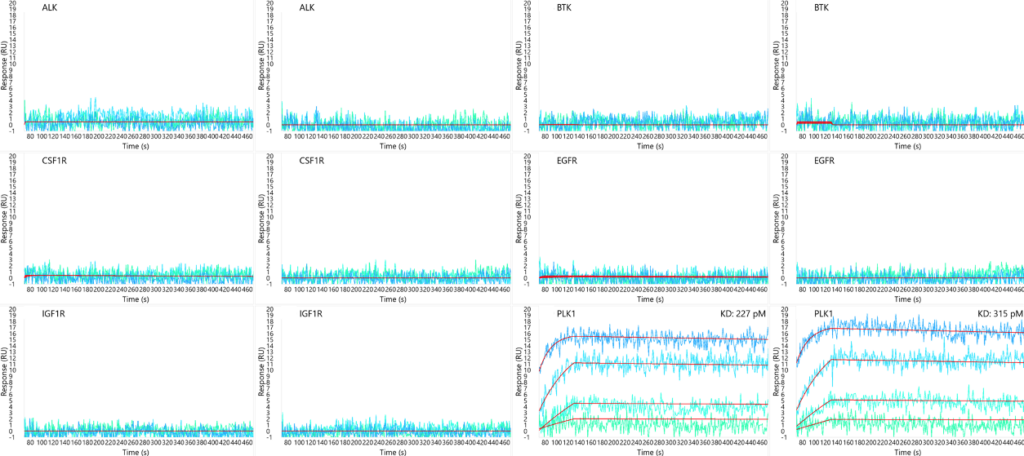
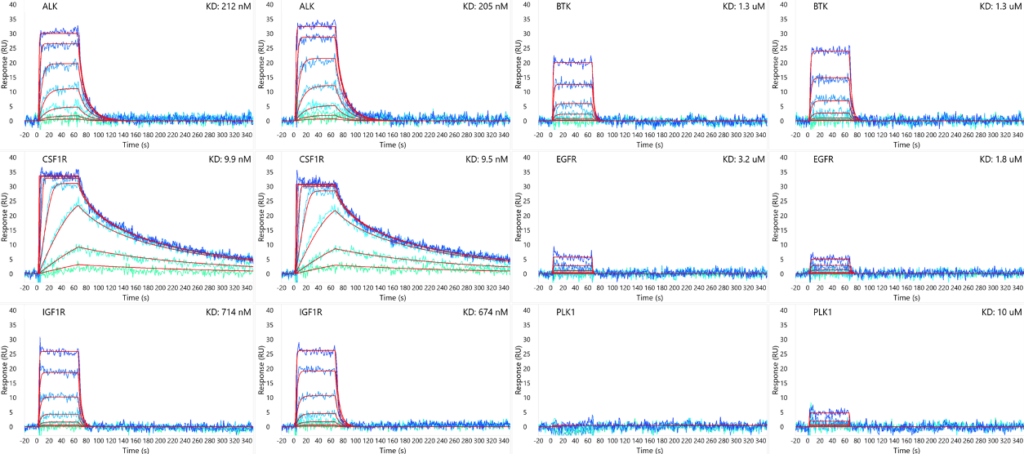
GSK-461364 binding to an array of kinases in duplicate. High selectivity is demonstrated for only PLK1 as expected.
