Key Benefits of Epitope Analysis Software
The Epitope software was designed to process the massive data sets created by the HT SPR technology from Carterra, where 384 ligands can be tested against 384 analytes. Prior to the development of this software, the data analysis of even small epitope binning panels could be a time intensive and laborious process.
Key Benefits:
- Rapidly and easily analyze and visualize up to 384×384 epitope binning studies.
- Quickly explore and curate the data. Plotting of injections, ligand spots, or other groups of interactions is possible from any point in the program, allowing users to view the raw data alongside the heat map generation, sorting and bin allocations.
- Easily define measurement points and generate a heat map showing blocking and sandwiching interactions.
- Sort the heat map into like-behaved antibodies and assign them to bins using custom algorithms and software automation.
Current Version: Epitope V.1.9.2.4505
Release Date: May 13, 2024
The latest version of Epitope offers a number of improvements, including:
- Application now manages .sprdata files
- Enhanced dendrogram color assignments and descriptions
- Bay and Position information added to Data tables
- All tables in Data tab mimic the copy and paste functions of Excel
Epitope Release Installation Notes
High Throughput Epitope Binning
A critical step in the discovery of biotherapeutics is to characterize and group a library of monoclonal antibodies by the epitope binding regions generated against a specific antigen. This “epitope binning” enables the maintenance of epitope diversity and provides important information to broaden intellectual property protection. The LSA delivers label-free, high throughput SPR that enables high throughput epitope binning of 384 antibodies directly from supernatant using less than 200 µL of each clone, overcoming the sample consumption, throughput limitations, and/or labeling restrictions of current methods. Carterra has developed a powerful integrated hardware and software platform that delivers the full potential of label-free, high throughput antibody binning.
What is Epitope Binning?
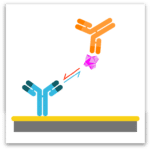
Premix Epitope Binning
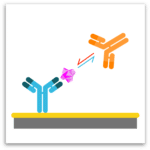
Classical Epitope Binning
In epitope binning, antibodies are tested in a pairwise combinatorial manner, and those that compete for the same binding region are grouped together into bins. In a classical epitope binning sandwich assay, a mAb analyte is tested for binding to antigen that is first captured via an immobilized mAb. In a premix epitope binning assay, the antigen is saturated with a mAb analyte in solution and this preformed antigen/mAb complex is tested for binding to an immobilized mAb; its binding response is compared with that for antigen alone.
Why Perform Epitope Binning?
In the quest for therapeutic monoclonal antibodies (mAbs), tools that allow an optimal candidate to be selected from a large number of leads can make the difference between a successful program and a clinical failure. A mAb’s epitope correlates with its functional activity, but the in silico prediction of B-cell epitopes is not yet possible, so epitope selection remains an empirical process. Organizing mAbs into epitope families or “bins” can be helpful, because mAbs that target similar epitopes often share a similar function. An epitope bin with functional activity can provide several potential leads to choose from. Conversely, if mAbs from multiple epitope bins exhibit functional activity, this implies different mechanisms of action, which can be advantageous when pursuing an oligoclonal therapy to treat some cancers or infectious diseases where simultaneous targeting of more than one biological pathway may be needed.
Commonly, however, antibody affinity remains the primary selection mechanism. By using affinity for selection, the result may be the selection of a panel of antibodies that bind to at most one or a few epitopes. This epitope bias can be problematic if the functional epitopes are not represented by those selected with the highest affinity.
Best Way to do Epitope Binning?
Previously, epitope binning has required significant time, sample and manpower for experimental setup and data analysis. This has limited its application to small numbers of samples later in the development process. Carterra’s LSA integrates a turnkey, high throughput SPR solution requiring minimal hands-on time, while delivering unmatched data from which to make informed selection decisions.
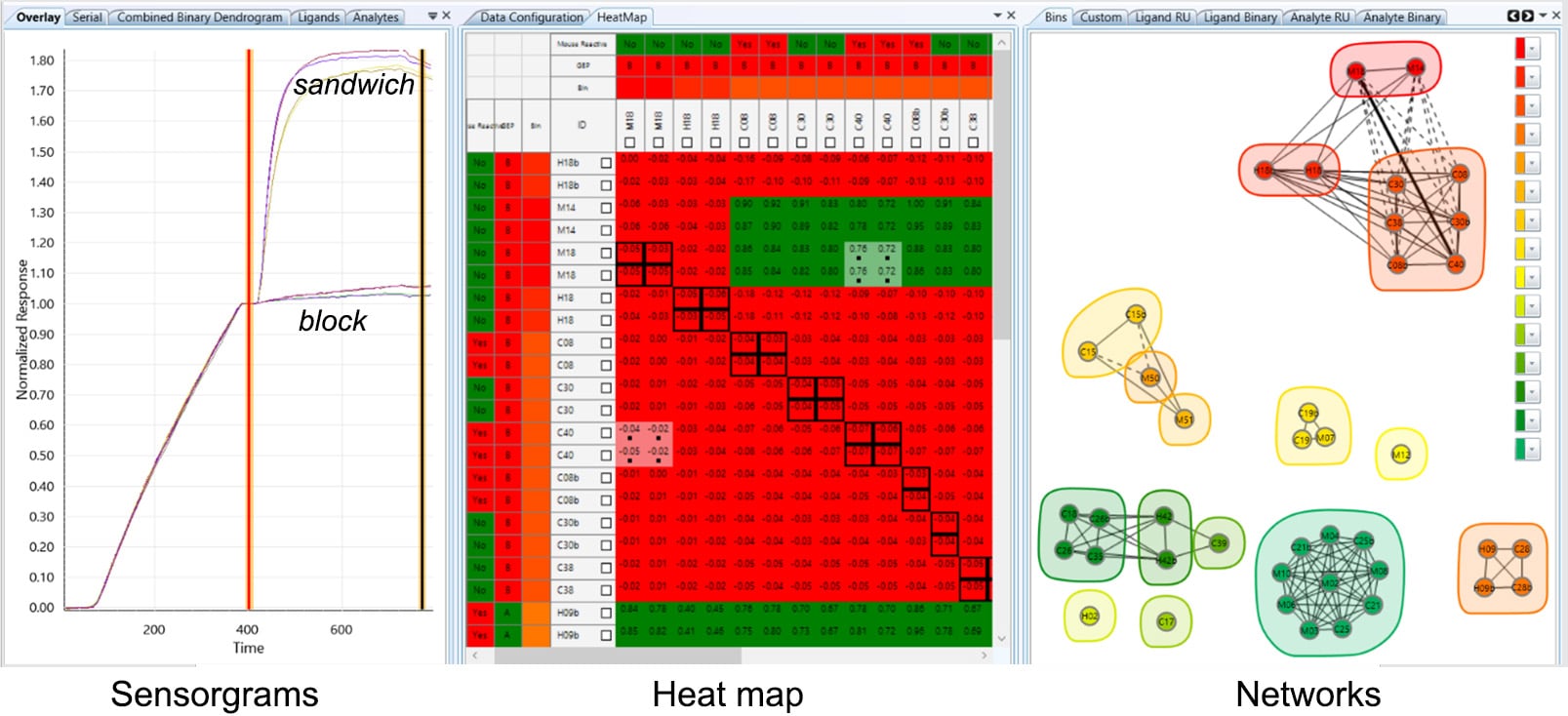
Data is linked across three visualization panels: sensorgrams, heat map and networks.
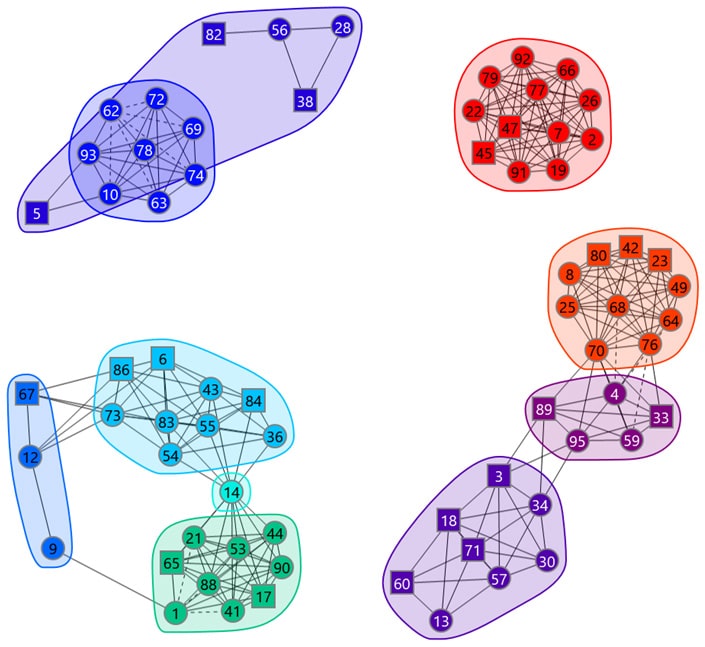
Our network plotting software provides an intuitive way of navigating the epitope binning results. In a network plot, the nodes represent the antibodies (identified by number), the chords indicate the pairwise blocking relationships (asymmetric blocks have dotted lines), and the envelopes inscribe the bins.
Minimum Hardware Specifications for Epitope Analysis Software
| Processor | |
| Minimum | 1.7 GHz Processor |
| Recommended | i9 Processor 3.6 GHz or higher |
| Operating System | |
| Minimum | Windows 10 |
| Data Connection | |
| Minimum | USB 2.0 when external drives are used |
| Recommended | USB 3.0 when external drives are used |
| Memory | |
| Minimum | 16 GB RAM |
| Recommended | 32 GB RAM |
| Graphics Display | |
| Minimum | 1280×1024 |
| Recommended | 3840×2160 |
| Software Installation and Activation | |
| Minimum | Internet connection required |
| Hard Disk | |
| Minimum | HDD with >10GB of free space |
| Recommended | SSD with >10GB of free space |
